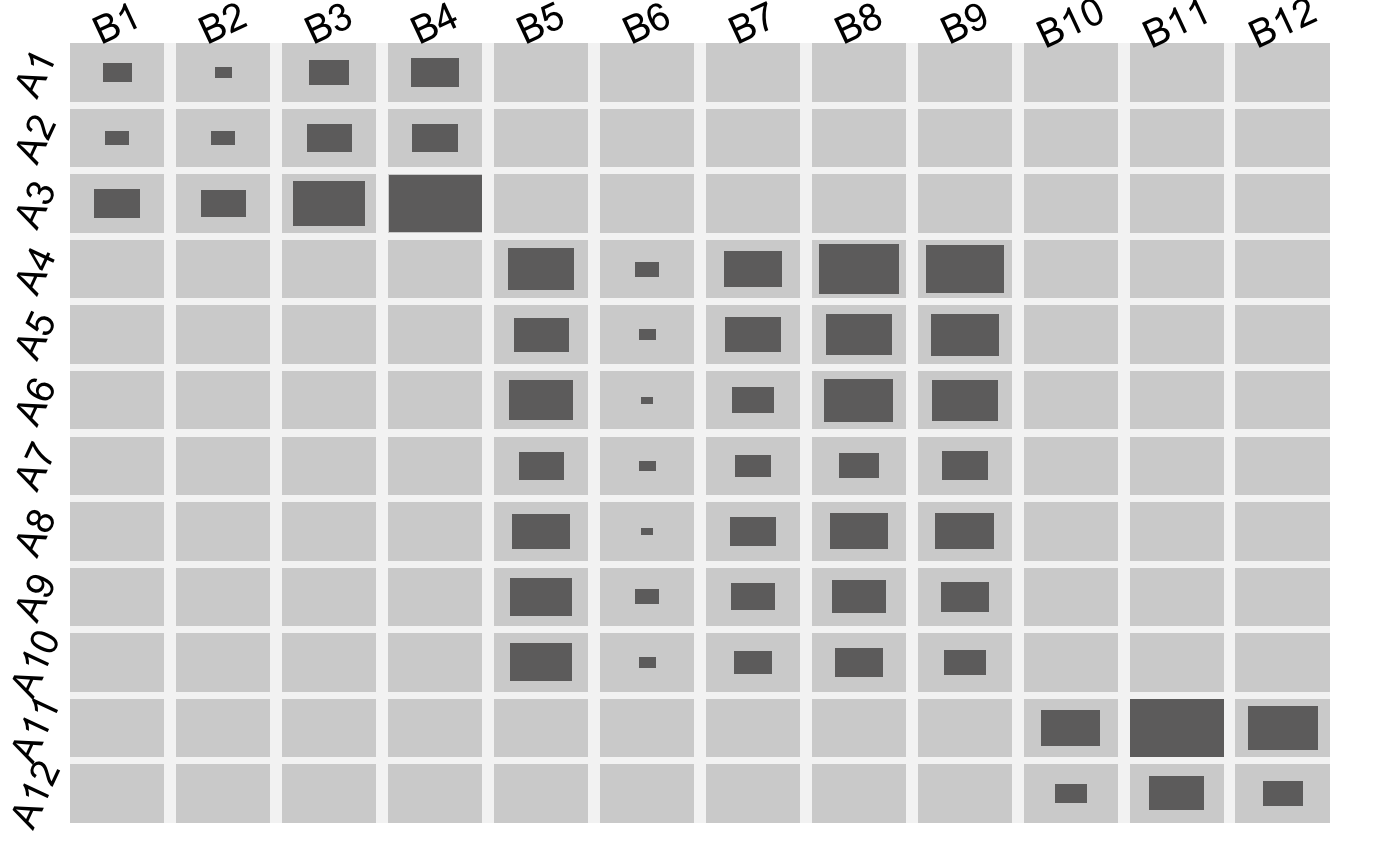
block-structured arrays
arsim.RdGenerates an array or matrix that includes k fully separated block-clusters.
arsim(n, dim, k, noise = 0, shuffle = TRUE, v = 0.1, minc = 1, exp.prop = NULL, min.prop = 1/dim/4, noise.type = "s", dimnames = list(LETTERS, 1:max(dim)))
Arguments
| n | The number of observations in the array. |
|---|---|
| dim | The dimension of the array. |
| k | The number of clusters. 1 for no clusters. |
| noise | The proportion of noise among the observations. There are two choices for |
| shuffle | Whether or not to shuffle the original category orders randomly. |
| v | A variability parameter for the assignment of the observations to the block clusters. Small values lea |
| minc | The minimum number of categories each cluster must have in each variable. E.g. |
| exp.prop | Optional: expected proportions of the observations which fall into the block clusters. |
| min.prop | Minimum proportion of observations in each cluster. |
| noise.type | Either |
| dimnames | A list of 2: The first entry defines the variable labels (default: A,B,C,...) and the second entry defines the category labels (default 1:k). |
Value
a simulated data array
Details
Not a very sophisticated way of generating random arrays but it serves for tests and illustrations of the other functions.
Examples
#> viewport[base]#> [[1]] #> [1] "a" "b" "c" "d" "e" "f" "g" "h" "i" "j" "k" "l" #> #> [[2]] #> [1] "a" "b" "c" "d" "e" "f" "g" "h" "i" "j" "k" "l" #>#> [[1]] #> [1] "0b" "0j" "0g" "0k" "0d" "0c" "0e" "0f" "0i" "0h" "0a" #> #> [[2]] #> [1] "1a" "1f" "1b" "1g" "1c" "1d" "1e" #> #> [[3]] #> [1] "2b" "2e" "2c" "2d" "2a" #># NOT RUN { A2<- arsim(1000, c(12,12,12), 3, shuffle = FALSE) fluctile3d(A2, shape ="oct") # }