class: center, middle, inverse, title-slide # Tidying and Joining Data with <code>tidyr</code> ## (Slides by Heike Hofmann) --- ## Recall - Sources of Messiness 1. Column headers are values, not variable names.<br> e.g. *treatmenta, treatmentb* 2. Multiple variables are stored in one column.<br> e.g. *Fall 2015, Spring 2016* or *"1301 8th St SE, Orange City, Iowa 51041 (42.99755, -96.04149)", "2102 Durant, Harlan, Iowa 51537 (41.65672, -95.33780)"* 3. Multiple observational units are stored in the same table. 4. A single observational unit is stored in multiple tables. --- ## Recall - Tidy data 1. Each variable forms one column. 2. Each observation forms one row. 3. Each type of observational unit forms a table. <!----- # Keys and Measurements ## Finding your keys - Example (1) 100 patients are randomly assigned to a treatment for heart attack, measured 5 different clinical outcomes. ## Finding your keys - Example (1) 100 patients are randomly assigned to a treatment for heart attack, measured 5 different clinical outcomes. - key: patient ID - factor variable (design): treatment - measured variables: 5 clinical outcomes ## Finding your keys - Example (2) Randomized complete block trial with four fields, four different types of fertilizer, over four years. Recorded total corn yield, and fertilizer run off ## Finding your keys - Example (2) Randomized complete block trial with four fields, four different types of fertilizer, over four years. Recorded total corn yield, and fertilizer run off - key: fields, types of fertilizer, year - measurement: total corn yield, fertilizer run off ## Finding your keys - Example (3) Cluster sample of twenty students in thirty different schools. For each school, recorded distance from ice rink. For each student, asked how often they go ice skating, and whether or not their parents like ice skating ## Finding your keys - Example (3) Cluster sample of twenty students in thirty different schools. For each school, recorded distance from ice rink. For each student, asked how often they go ice skating, and whether or not their parents like ice skating - key: student ID, school ID - measurement: distance to rink, #times ice skating, parents' preference ## Finding your keys - Example (4) For each person, recorded age, sex, height and target weight, and then at multiple times recorded their weight ## Finding your keys - Example (4) For each person, recorded age, sex, height and target weight, and then at multiple times recorded their weight - key: *patient ID*, date - measurement: *age, sex, height, target weight*, current weight *only patient ID is needed for variables in italics* ---> --- ## Messy (3) Messy (3): *Multiple observational units are stored in the same table.* What does that mean? The *key is split*, i.e. for some values all key variables are necessary, while other values only need some key variables. 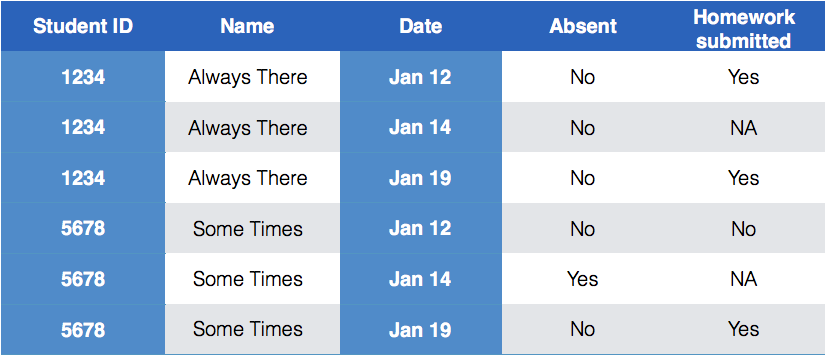 --- ## Why do we need to take care of split keys? - Data redundancy introduces potential problems (same student *should* have the same student ID) - to check data consistency, we split data set into parts - this process is called *normalizing* - normalization reduces overall data size - useful way of thinking about objects under study --- ## Tidying Messy (3) Splitting into separate datasets: 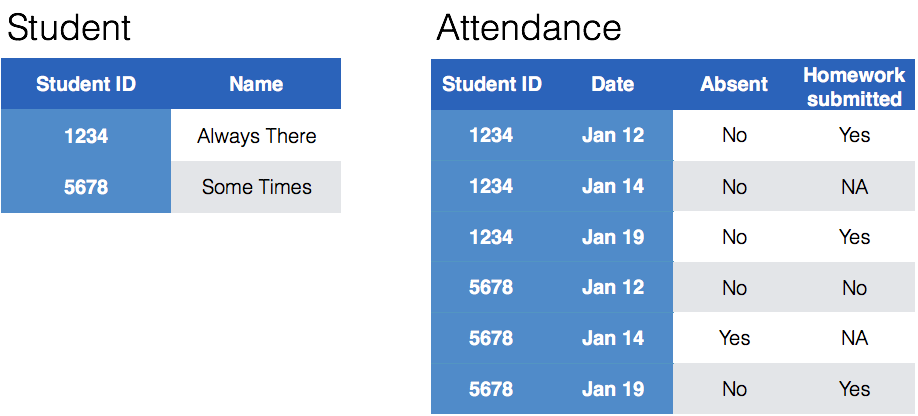 --- class: inverse, middle, center # Messy (4) --- ## Messy (4) **Messy (4)**: Values for a single observational unit are stored across multiple tables. After data are normalized by splitting, we want to de-normalize again by **joining** datasets. --- ## Example: Lahman package Sean Lahman is a database journalist, who started databases of historical sports statistics, in particular, the Lahman database on baseball. ```r library(Lahman) LahmanData ``` ``` ## file class nobs nvar title ## 1 AllstarFull data.frame 4993 8 AllstarFull table ## 2 Appearances data.frame 99466 21 Appearances table ## 3 AwardsManagers data.frame 171 6 AwardsManagers table ## 4 AwardsPlayers data.frame 6026 6 AwardsPlayers table ## 5 AwardsShareManagers data.frame 401 7 AwardsShareManagers table ## 6 AwardsSharePlayers data.frame 6705 7 AwardsSharePlayers table ## 7 Batting data.frame 99846 22 Batting table ## 8 BattingPost data.frame 11294 22 BattingPost table ## 9 CollegePlaying data.frame 17350 3 CollegePlaying table ## 10 Fielding data.frame 167938 18 Fielding table ## 11 FieldingOF data.frame 12028 6 FieldingOF table ## 12 FieldingPost data.frame 11924 17 FieldingPost data ## 13 HallOfFame data.frame 4088 9 Hall of Fame Voting Data ## 14 Managers data.frame 3370 10 Managers table ## 15 ManagersHalf data.frame 93 10 ManagersHalf table ## 16 Master data.frame 18589 26 Master table ## 17 Pitching data.frame 43330 30 Pitching table ## 18 PitchingPost data.frame 4945 30 PitchingPost table ## 19 Salaries data.frame 24758 5 Salaries table ## 20 Schools data.frame 1207 5 Schools table ## 21 SeriesPost data.frame 298 9 SeriesPost table ## 22 Teams data.frame 2775 48 Teams table ## 23 TeamsFranchises data.frame 120 4 TeamFranchises table ## 24 TeamsHalf data.frame 52 10 TeamsHalf table ``` --- ## Lahman database The Lahman database consists of 24 data frames that are linked by `playerID`. This is clean, but not very readable. <br> The `Master` table includes names and other attributes for each player. <br> **Joining** multiple tables helps us to bring together (pieces of) information from multiple sources. --- ## Example: Hall of Fame ```r HallOfFame <- HallOfFame %>% group_by(playerID) %>% mutate(times = order(yearID)) HallOfFame %>% ggplot(aes(x = yearID, y = votes/needed, colour = inducted)) + geom_hline(yintercept = 1, colour = "grey20", size = .1) + geom_line(aes(group = playerID), colour = "black", size = 0.2) + geom_point() ``` 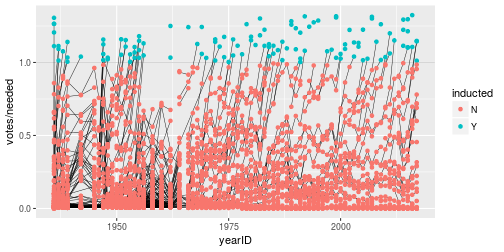<!-- --> --- ## Hall of Fame - how many attempts? We'd like to label all the last attempts - and not just with the `playerID` ```r HallOfFame %>% ggplot(aes(x = times, y = votes/needed, colour = inducted)) + geom_hline(yintercept = 1, colour = "grey20", size = .1) + geom_line(aes(group = playerID), colour = "black", size = 0.2) + geom_point() ``` 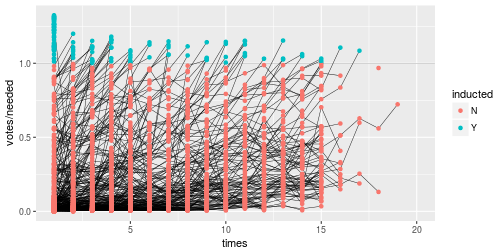<!-- --> --- ## Joins - general idea 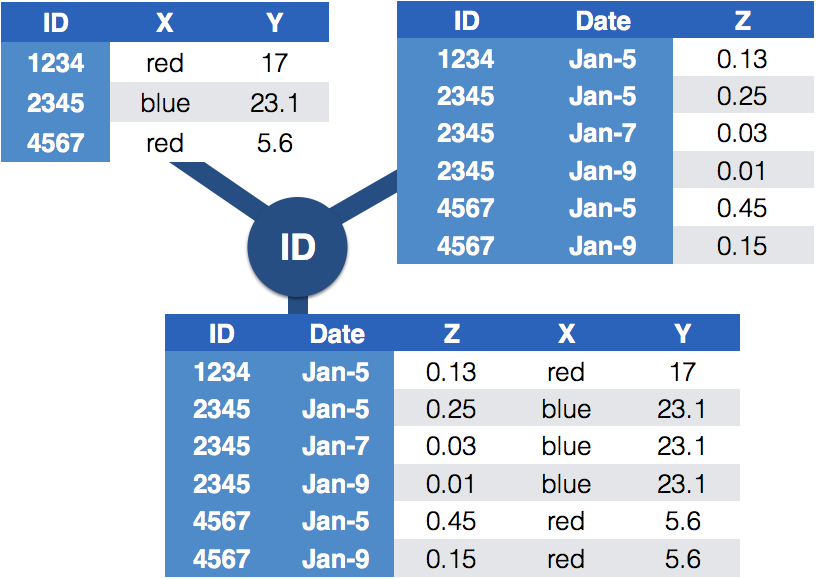 --- ## Joins - more specific idea - Data sets are joined along values of variables. - In `dplyr` there are various join functions: `left_join`, `inner_join`, `full_join`, ... - Differences between join functions are only visible if not all values in one set have values in the other --- ## Simple example data ```r df1 <- data.frame(id = 1:6, trt = rep(c("A", "B", "C"), rep=c(2,1,3)), value = c(5,3,7,1,2,3)) df1 ``` ``` ## id trt value ## 1 1 A 5 ## 2 2 B 3 ## 3 3 C 7 ## 4 4 A 1 ## 5 5 B 2 ## 6 6 C 3 ``` ```r df2 <- data.frame(id=c(4,4,5,5,7,7), stress=rep(c(0,1), 3), bpm = c(65, 125, 74, 136, 48, 110)) df2 ``` ``` ## id stress bpm ## 1 4 0 65 ## 2 4 1 125 ## 3 5 0 74 ## 4 5 1 136 ## 5 7 0 48 ## 6 7 1 110 ``` --- ## Left join - all elements in the *left* data set are kept - non-matches are filled in by `NA` - `right_join` works symmetric ```r left_join(df1, df2, by="id") ``` ``` ## id trt value stress bpm ## 1 1 A 5 NA NA ## 2 2 B 3 NA NA ## 3 3 C 7 NA NA ## 4 4 A 1 0 65 ## 5 4 A 1 1 125 ## 6 5 B 2 0 74 ## 7 5 B 2 1 136 ## 8 6 C 3 NA NA ``` --- ## Inner join - only matches from both data sets are kept ```r inner_join(df1, df2, by = "id") ``` ``` ## id trt value stress bpm ## 1 4 A 1 0 65 ## 2 4 A 1 1 125 ## 3 5 B 2 0 74 ## 4 5 B 2 1 136 ``` --- ## Full join - all ids are kept, missings are filled in with `NA` ```r full_join(df1, df2, by = "id") ``` ``` ## id trt value stress bpm ## 1 1 A 5 NA NA ## 2 2 B 3 NA NA ## 3 3 C 7 NA NA ## 4 4 A 1 0 65 ## 5 4 A 1 1 125 ## 6 5 B 2 0 74 ## 7 5 B 2 1 136 ## 8 6 C 3 NA NA ## 9 7 <NA> NA 0 48 ## 10 7 <NA> NA 1 110 ``` --- ## Traps of joins - sometimes we unexpectedly cannot match values: missing values, different spelling, ... - join can be along multiple variables, e.g. `by = c("ID", "Date")` - joining variable(s) can have different names, e.g. `by = c("State" = "Name")` - always make sure to check dimensions of data before and after a join - check on missing values; help with that: `anti_join` --- ## Anti join - a neat function in `dplyr` - careful, not symmetric! ```r anti_join(df1, df2, by="id") # no values for id in df2 ``` ``` ## id trt value ## 1 1 A 5 ## 2 2 B 3 ## 3 3 C 7 ## 4 6 C 3 ``` ```r anti_join(df2, df1, by="id") # no values for id in df1 ``` ``` ## id stress bpm ## 1 7 0 48 ## 2 7 1 110 ``` --- ## Joining baseball data Does lifetime batting average make a difference in a player being inducted? ```r Batting2 <- Batting %>% group_by(playerID) %>% mutate(BatAvg = H/AB) %>% summarise(LifeBA = mean(BatAvg, na.rm=TRUE)) hof_bats <- inner_join(HallOfFame %>% filter(category == "Player"), Batting2, by = c("playerID")) hof_bats %>% ggplot(aes(x = yearID, y = LifeBA, group = playerID)) + geom_point(aes(color = inducted)) ``` 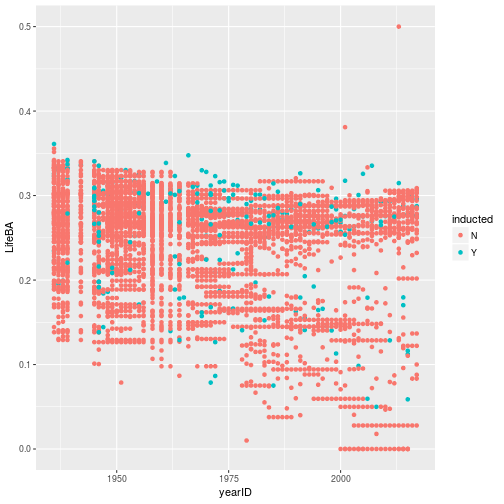<!-- --> --- ## Joining Baseball Data (2/2) What about pitchers? Are pitchers with lower lifetime ERAs more likely to be inducted? ```r Pitching2 <- Pitching %>% group_by(playerID) %>% summarise(LifeERA = mean(ERA, na.rm = TRUE)) hof_pitch <- inner_join(HallOfFame %>% filter(category == "Player"), Pitching2, by = c("playerID")) hof_pitch %>% ggplot(aes(x = yearID, y = LifeERA, group = playerID)) + geom_point(aes(color = inducted)) ``` ``` ## Warning: Removed 2 rows containing missing values (geom_point). ``` 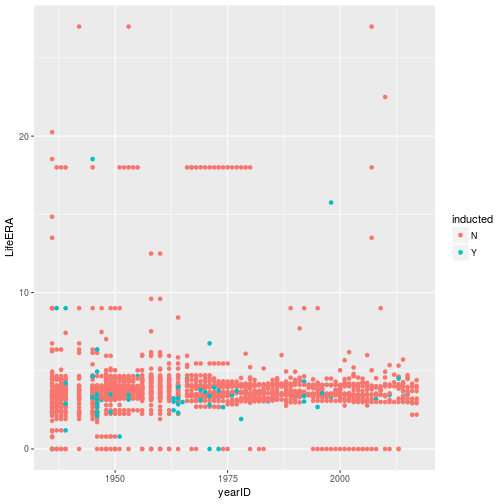<!-- --> --- class: inverse ## Your turn - Load the `Lahman` package into your R session. - Join (relevant pieces of) the `Master` data set and the `HallOfFame` data. - For the `ggplot2` chart label all last attempts of individuals with 15 or more attempts. Make sure to deal with missing values appropriately. <!--- ## Chart of induction ```r Voted %>% ggplot(aes(x = attempt, y = votes/needed)) + geom_hline(yintercept = 1, colour = "grey25", size = 0.1) + geom_line(aes(group = playerID), colour = "grey35", size = 0.2) + geom_point(aes(colour = inducted)) ``` ## Getting the dataset for the labels ```r labels <- Voted %>% group_by(playerID) %>% summarize( votes = votes[which.max(attempt)], needed = needed[which.max(attempt)], attempt = max(attempt), name = paste(unique(nameFirst), unique(nameLast)) ) ``` --->