Convert sequence data to a format suitable for logo plots
Usage
ggfortify(
data,
sequences,
treatment = NULL,
weight = NULL,
method = "shannon",
missing_encode = c(".")
)Arguments
- data
data frame with the sequences
- sequences
variable containing the sequences
- treatment
co-variate(s) used in collecting sequence data
- weight
numeric variable of weights
- method
either "shannon" or "frequency" for Shannon information or relative frequency of element by position.
- missing_encode
which character value(s) signifies a missing element in a sequence? defaults to `.`.
Examples
# \donttest{
library(ggplot2)
data(sequences)
ggplot(data = ggfortify(sequences, peptide, treatment = class)) +
geom_logo(aes(x = class, y = bits, fill = Water, label = element)) +
facet_wrap(~position)
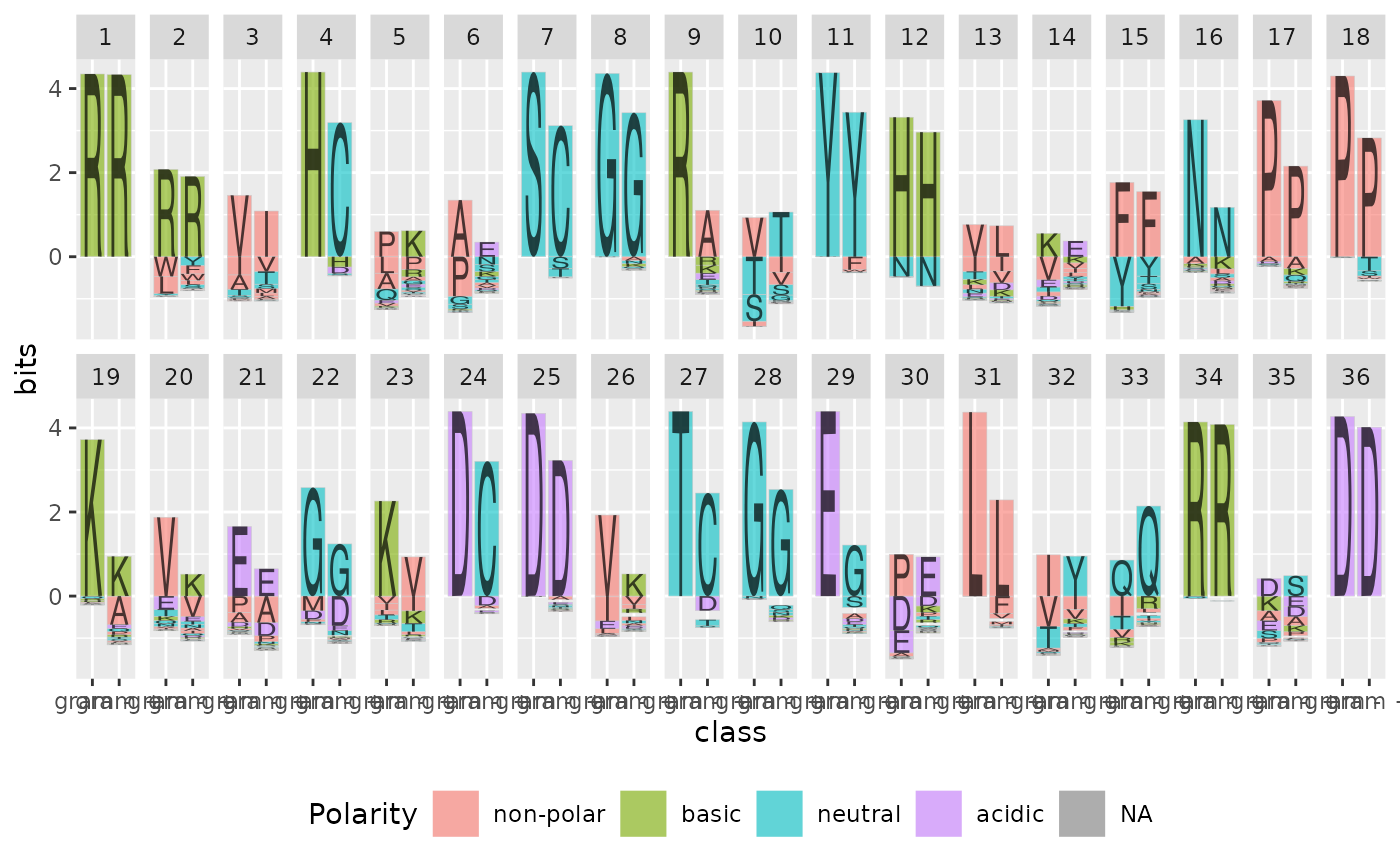
 ggplot(data = ggfortify(sequences, peptide, treatment = class)) +
geom_logo(aes(x = class, y = bits, fill = Polarity, label = element)) +
facet_wrap(~position, ncol = 18) +
theme(legend.position = "bottom")
ggplot(data = ggfortify(sequences, peptide, treatment = class)) +
geom_logo(aes(x = class, y = bits, fill = Polarity, label = element)) +
facet_wrap(~position, ncol = 18) +
theme(legend.position = "bottom")
 # }
# }